Introduction¶
The idea behind Cloudgene is quite simple: If you are able to execute your program or Hadoop application on the command line, take some minutes and write a YAML configuration to connect your program with Cloudgene. Doing so, you are able to transform your Hadoop command line program (or a set of programs) into a web-based service, present your collaborators a scalable best practices workflow and provide reproducible science.
Project structure¶
First of all, we create a new project folder for our new application. This folder must contain a file called cloudgene.yaml. This file is also called the Manifest File and includes all information needed in order to install and execute your programs or pipelines. This folder contains also all other files that are needed by your workflow steps. For example binaries, a jar file of a ready-to-use MapReduce program, a PIG script or some R-Markdown script.
A basic Cloudgene application usually looks something like this:
my-cloudgene-app ├── cloudgene.yaml ├── sample-mapredduce-program.jar ├── sample-rmarkdown-report.Rmd ├── some-folder | ├── dataset2.csv | └── my-binary └── README.md
After we are finished developing our application, we create a zip file of this folder. This zip file contains all needed files and can be deployed to any Cloudgene webserver by uploading it.
Read more about installing applications.
cloudgene.yaml¶
The file content starts with a simple header containing general information about the application, followed by the description of input/output parameters as well as all steps of the workflows:
A simple example looks like this:
id: tool-name name: Tool Name description: tool-description category: tool-category version: 1.0 website: http://www.my-website.com
The two most important fields are id, name and version, without them your application won’t be able to install. The id and version fields are used together to create a unique id. description and website should be used to describe your application.
The next step is to add the workflow section to your configuration file to define steps as well as input- and output-parameters.
Defining steps¶
The simplest way to model a workflow is to create a list of steps where each step depends on its forerunner. Steps are defined in the steps section where each step is defined by a name and type specific properties.
A simple example with two steps looks like this:
id: hello-cloudgene name: Hello Cloudgene version: 1.0 workflow: steps: - name: Step1 cmd: /bin/echo hey cloudgene developer! I am step 1. stdout: true - name: Step2 cmd: /bin/echo hey cloudgene developer! I am step 2. stdout: true
In this example we used the command line tool echo to print out some text. However, Cloudgene supports a variety of different step types which can be combined into one workflow to take advantage of different technologies.
To test our workflow we copy the content into a file named hello-cloudgene.yaml. Next, we can upload the zip file of our application to a Cloudgene webserver or we execute it on our developement machine with the cloudgene-cli program:
cloudgene run hello-cloudgene.yaml
Cloudgene 1.30.6 http://www.cloudgene.io (c) 2009-2018 Lukas Forer and Sebastian Schoenherr Built by lukas on 2018-11-19T10:14:17Z hello-cloudgene 1.0 [INFO] No external Haddop cluster set. [WARN] Cluster seems unreachable. Hadoop support disabled. [INFO] Submit job job-20181119-112052... [OUT] hey cloudgene developer! I am step 1. [OK] Execution successful. [OUT] hey cloudgene developer! I am step 2. [OK] Execution successful. Done! Executed without errors. Results can be found in file:///home/lukas/new-cloudgene/job-20181119-112052
We see that Cloudgene executes our workflow and prints the text to the terminal.
Defining input parameters¶
Input parameters are defined in the inputs section where each parameter is defined by an unique id, a textual description and a type.
We extend the example above by an input parameter to set the message:
id: hello-cloudgene name: Hello Cloudgene version: 1.0 workflow: steps: - name: Step1 cmd: /bin/echo hey cloudgene developer! $message stdout: true inputs: - id: message description: Message type: text
If the workflow is executed on a Cloudgene Webserver, a web-interface is automatically created where the user has to enter the message. However, if we execute the workflow using cloudgene-cli, the parameter has to be set as a command-line argument:
cloudgene run hello-cloudgene.yaml --message "Using an input parameter is easy!"
Cloudgene 1.30.6 http://www.cloudgene.io (c) 2009-2018 Lukas Forer and Sebastian Schoenherr Built by lukas on 2018-11-19T10:14:17Z hello-cloudgene 1.0 [INFO] No external Haddop cluster set. [WARN] Cluster seems unreachable. Hadoop support disabled. [INFO] Submit job job-20181119-112527... Input values: message: Using an input parameter is easy! [OUT] hey cloudgene developer! Using an input parameter is easy! [OK] Execution successful. Done! Executed without errors. Results can be found in file:///home/lukas/new-cloudgene/job-20181119-112527
Serve the webservice¶
Before you can start a webservice for your workflow, you have to install it:
cloudgene install hello-cloudgene.yaml
Cloudgene 1.30.6 http://www.cloudgene.io (c) 2009-2018 Lukas Forer and Sebastian Schoenherr Built by lukas on 2018-11-19T10:14:17Z Installing application hello-cloudgene... Process file hello-cloudgene.yaml.... [OK] 1 Applications installed: APPLICATION VERSION STATUS FILENAME hello-cloudgene 1.0 OK hello-cloudgene.yaml
Next, we can start the webserver:
cloudgene server
The webservice is available on http://localhost:8082. Please use username admin and password admin1978 to login.
Click on Run and slect your application (hello-cloudgene). Cloudgene creates automatically a web-interface for your input parameters:
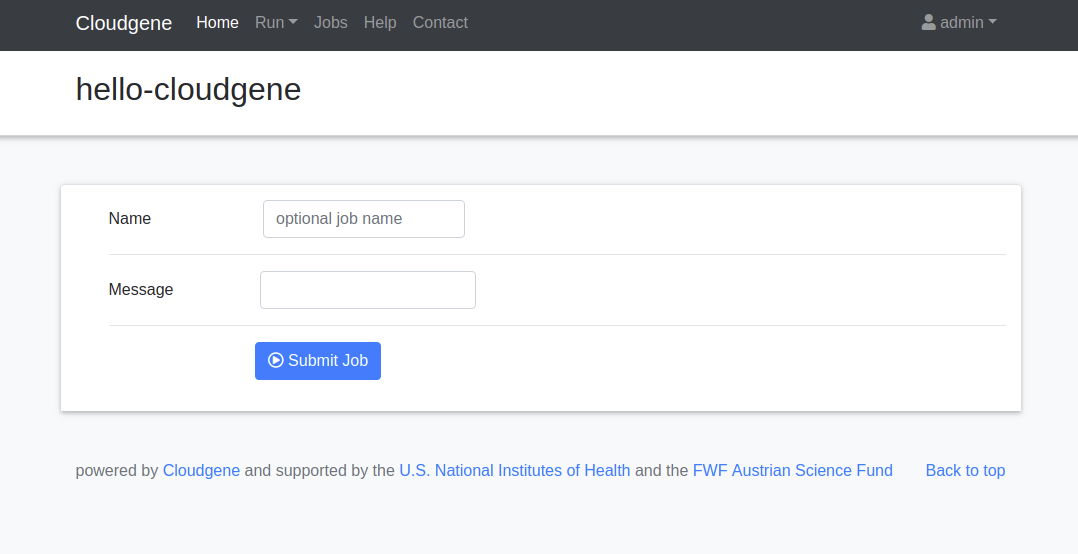
Enter a message and click on Submit Job to run your workflow. You should see the following job output:
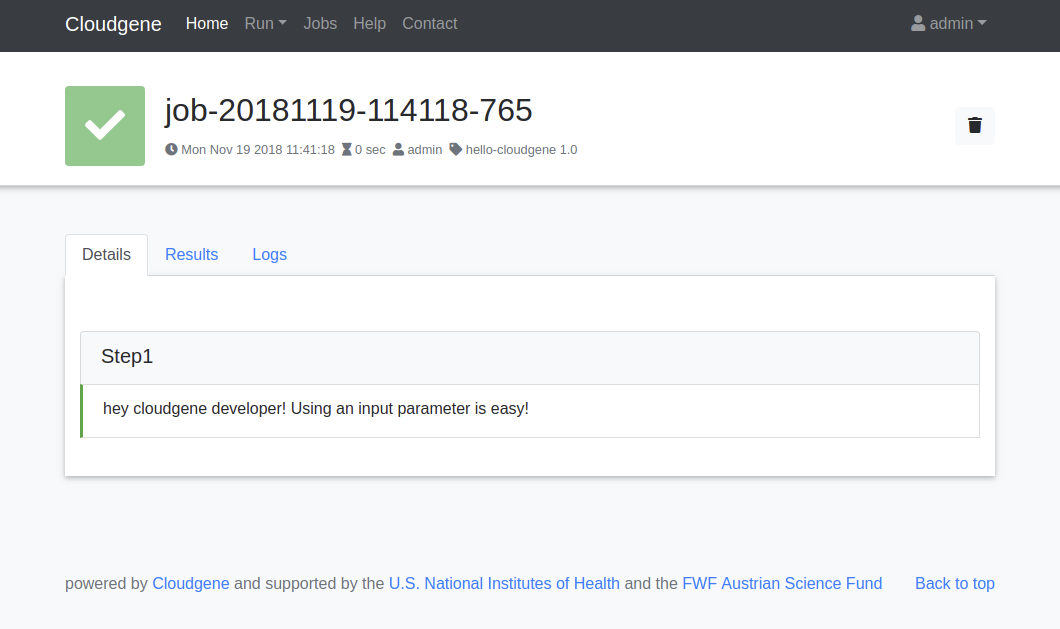
You can install different workflows in the same instance and define in the Admin Dashboard who has access to each of them.